gDR is an umbrella package for the gDR programmatic R interface.
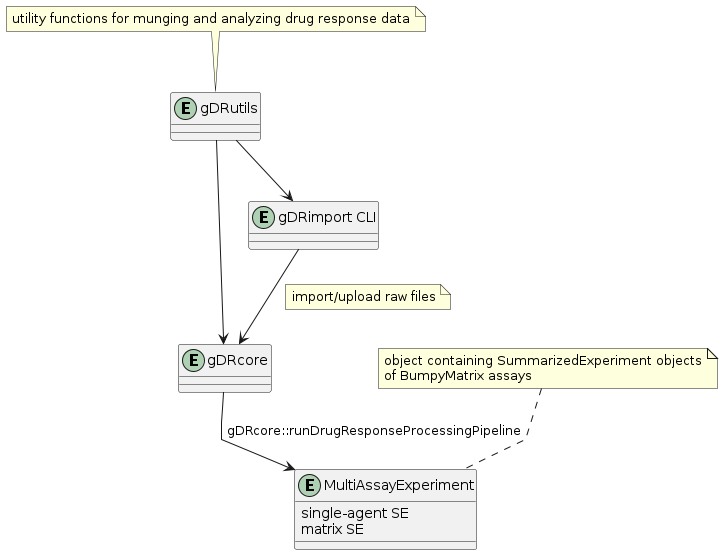
The gDR suite offers a full stack solution for processing drug response data. This enables a range of users across computational savvy, (i.e. lab scientists and computational scientists alike) to access the same, standardized data. The suite is made up of several core R packages.
Installation
The easiest way to use functions from gDR package is by using the Bioconductor.
- Install BiocManager
install.packages("BiocManager")- Install gDR packages
BiocManager::install(c("gDRstyle", "gDRtestData", "gDR"), version = "devel")You’ve just successfully set up the environment for the gDR umbrella package. Please see gDR vignette for examples of usage.
How to contribute
We are eager to make this tool useful for the community. We welcome any suggestions via GitHub issues, or pull requests for bugfixes/new features. When making pull requests, please first have a look at the style conventions in gDRstyle.